Optimizing the Performance of the T Cell Academy
Written by: Alexander Kirchmair and Otto Paans
As discussed in the previous blogs, in the INCITE project we are building an artificial immune niche, an academy for T cells where they learn the best ways to attack cancer cells. But how do we know what this academy should look like?
To explore this, we take inspiration from nature and analyze the immune niches that already exist in our bodies: the lymph nodes which are distributed throughout the human body. This is where immune responses are initiated. This happens through the education of T cells, which are taught how to recognize and eliminate a particular threat. If we want to understand this process, we have to investigate the interactions between the components of the lymph node:
- Fibroblasts, which are the structural cells of the lymph node and determine the intricate architecture of the T cell “classrooms”.
- Dendritic cells, which are the teachers of the T cells and activate them if needed.
- T cells, which only pass by briefly if dendritic cells don’t indicate that they have to stay for an “education” process.
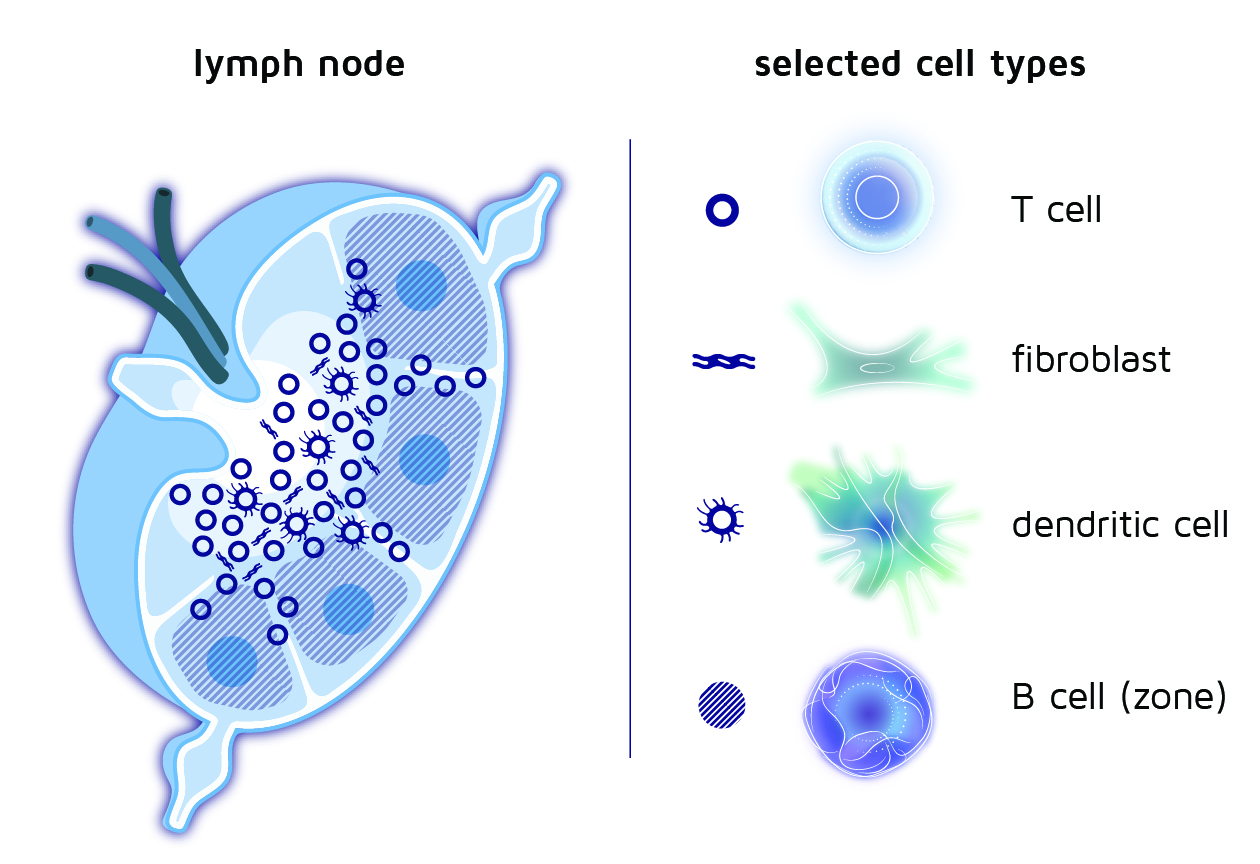
Figure 1: Schematic representation of a lymph node and approximate locations of various cell types
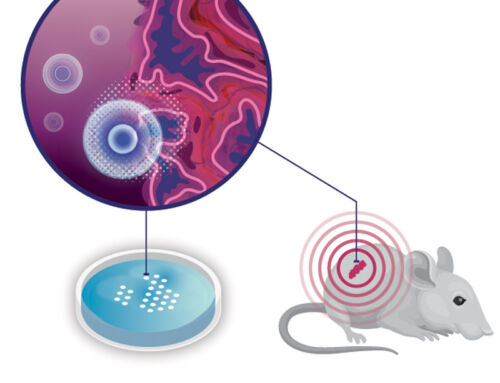
The preceding blog articles described how we build an artificial immune niche by combining 3D-printed and biological components into a more complex system, going through the components step by step. In this blog, we start with the complete system and try to break down how it works. To avoid getting lost in details, we compare lymph nodes that generate effective anti-tumor T cells to those that fail to do so, and then focus on the differences. After all, we would like to know what makes an artificial immune niche effective. If we can define the properties that makes a given immune niche effective, we can attempt to replicate its features. As we cannot study this in humans, we use mice that develop cancer and research lymph nodes that are close to a tumor. By comparing mice that successfully fight cancer with those that do not, we can identify the properties of the lymph node niche that result in effective anti-tumor immune responses.
As we want to obtain a complete picture of these differences, we aim for a comprehensive characterization of the lymph node cells.
So, first we must consider how cells differ from each other. All cells have the same genes, but some of them are turned on only in special cell types. For example, only killer T cells are able to turn on genes that produce substances making cancer cells commit suicide.
Memory T cells, by contrast, turn on genes that maintain their long-term health and longevity. Dendritic cells use even other genes to collect signals from their surroundings, enabling them to instruct T cells on which genes they should activate. So, measuring which genes are turned on or off in every cell within a lymph node lets us infer which cell types are present and what their function might be.
We can measure the activities of genes as follows. An activated gene produces temporary working copies of itself called transcripts. These transcripts contain a barcode specific to each gene. By reading this barcode using RNA sequencing, we can count the number of transcripts coming from each gene, providing a quantitative measure for their activation.
Transcriptomics is the methodology of measuring the transcripts in a cell, and we can use it for tens of thousands of genes at once.
Commonly, transcriptomics is carried out by isolating thousands to millions of cells from a sample and mixing them all together to obtain enough material for measurements. This is useful when samples consist of a homogeneous population of cells, and we used this to obtain detailed data on the differences in gene activation in T cells trained in vitro. However, these population-averaged data only provide insufficient information on the diversity of cell types found in lymph nodes in vivo. State-of-the-art methods also allow us to perform technically more challenging single-cell transcriptomics measurements to study the activity of genes in each of the individual cells in a sample.
A good way to think of this is to compare it with making a fruit smoothie. If we blend a fixed number of fruit types (let’s say, apples, lemons and raspberries) in a smoothie, we get a homogenous mix. From the smoothie, it is impossible to distinguish how many pieces of each fruit went into it, or to determine with certainty where each piece of fruit ends up. However, the smoothie has a certain taste and consistency that we can recognize. Likewise, bulk transcriptomic measurement allows us for creating a generalized picture of a population of given cells. But this only works because the population is homogenous – i.e. generated under artificial conditions.
But the cell population in a natural lymph node is not nearly as homogenous. And so, we acquire more insight if we analyze it piece by piece, using single-cell transcriptomic measurement. We are now as it were concerned with examining how many types of fruit there are, and how many of each we can identify. From this list, we acquire a far more precise picture.
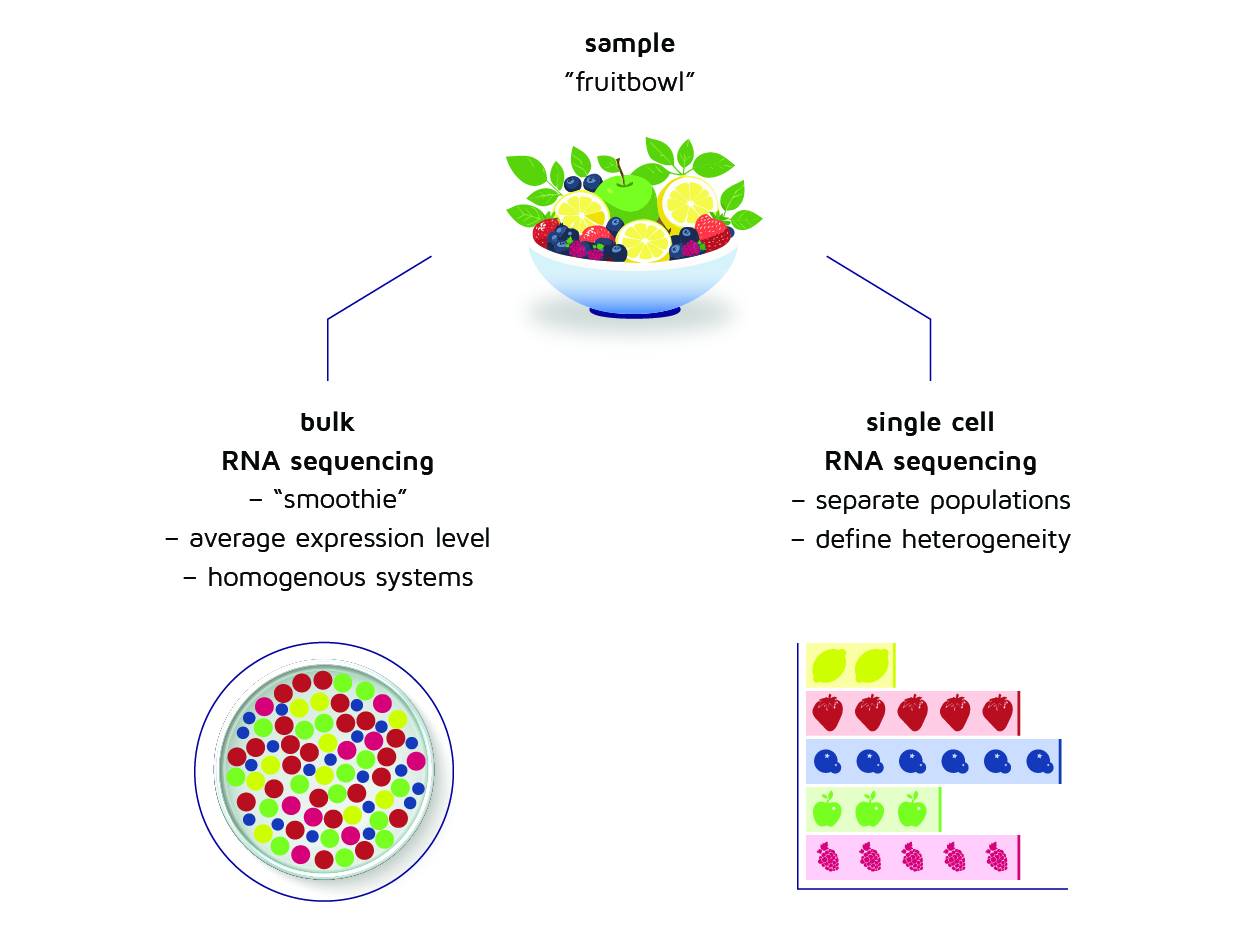
Figure 2: Differences between bulk RNA sequencing (i.e. “the smoothie”) versus single cell RNA sequencing (i.e. “the list”)
Single-cell transcriptomics enables us to computationally group cells by their gene activity profiles, so that we can infer which cell types and subtypes are present, and in what numbers. We can as it were define “populations” of cell types. By profiling the transcriptomes of tens of thousands of cells isolated from lymph nodes, we can identify the cells that lead to better or weaker anti-tumor immune responses.
Some subtypes of lymph node fibroblasts seem to be particularly good at generating the proper “classrooms” for T cell activation, making those the desired fibroblasts for the artificial immune niche. By contrast, we found that dendritic cells can comprise both stimulating and suppressive subtypes, so we need to take care in selecting the right teachers for anti-tumor T cell activation. You could say that we have identified “good” and “bad” teachers, so the challenge becomes to retain the good ones in the T cell academy.
Figure 3: Visual representation of various cell populations in a sample, each represented by a different colour (generic sample).
Apart from these features, there’s a lot more we can learn from the single-cell transcriptome data. If cells are stimulated by certain signals, they activate a defined set of genes. So, in turn, from the activated genes, we can reverse engineer which signals are present in the lymph nodes, and how they contribute to the teaching of different cell subtypes. That way, we can elucidate in detail which signals are needed to educate effective anti-tumor T cells.
Moreover, by specifically looking at genes that are known to transfer signals between cells (e.g., cytokines), or that are involved in the physical attachments between cells, we can reconstruct the major communication channels between the different cell subtypes.
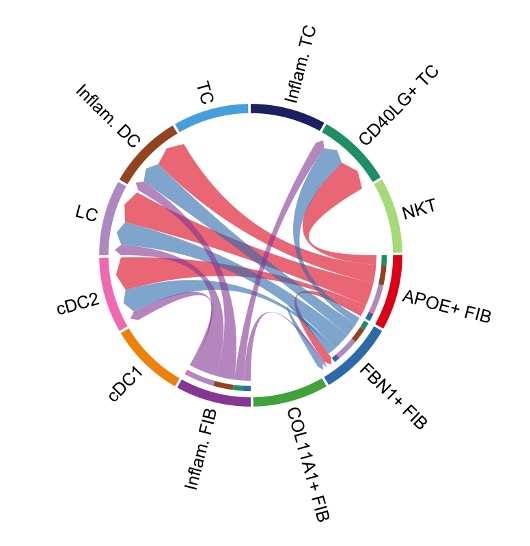
Figure 4: Cell-cell interaction map that shows the communication channels. Example from https://htmlpreview.github.io/?https://github.com/jinworks/CellChat/blob/master/tutorial/CellChat-vignette.html
Thereby, we find that fibroblasts interact with multiple T cell and dendritic cell subtypes in the lymph node, underlining their role as a critical component of the immune niche. Once we understand the details of these communication channels, we can use this knowledge to improve the design of the artificial niche by enable a more efficient flow of these signals, or by facilitating the physical attachments between the right cell types.
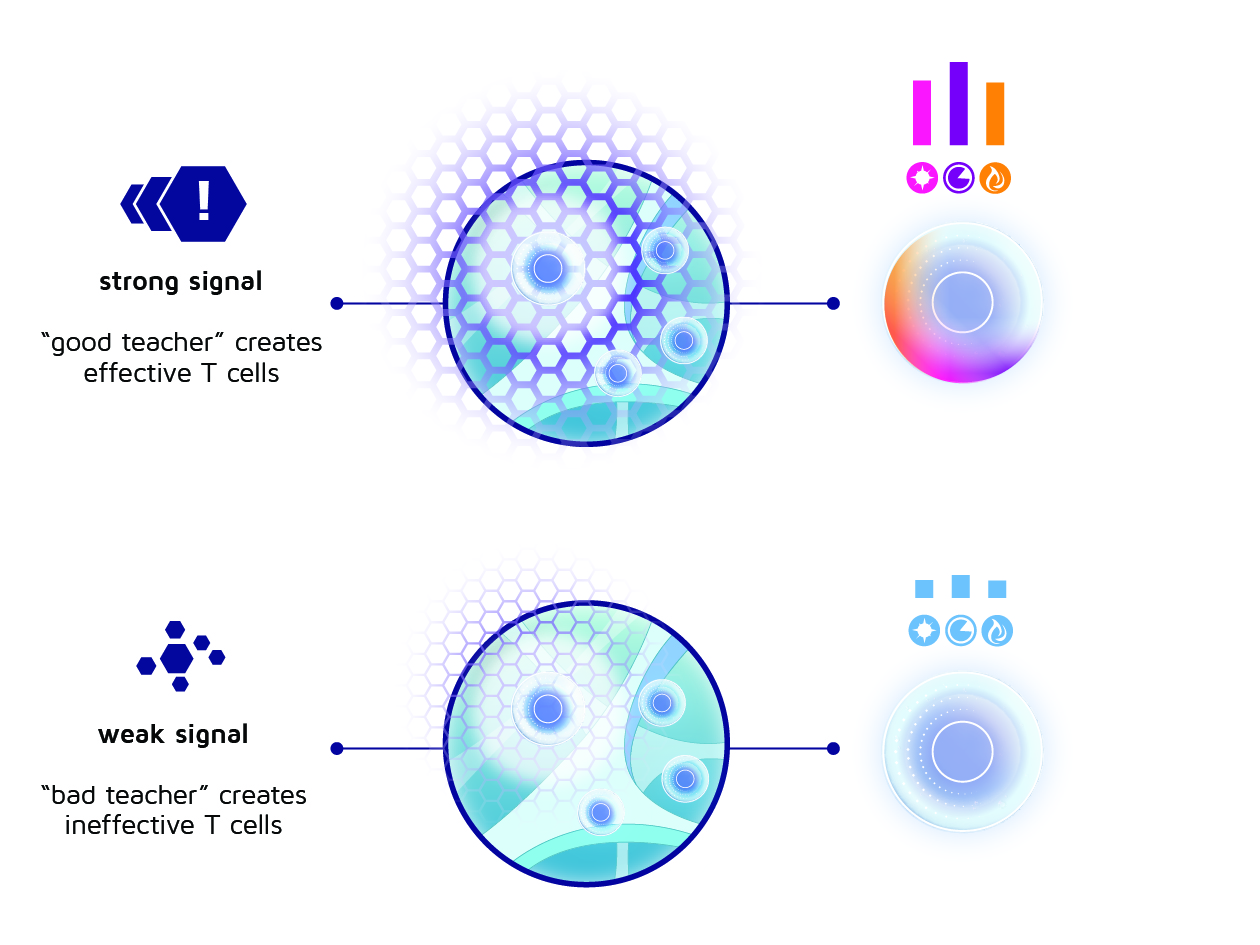
Figure 5: One of the INCITE aims: to select cells that possess strong signalling capabilties in order to impart their effectiveness to a next generation of T cells.
Overall, this gives us some hints about how we can generate the most effective artificial immune niche: First, we have to add all the “good” teachers, i.e., the right subtypes of fibroblasts and dendritic cells, and make sure they can properly grow and stay healthy within the niche. Second, we have to design the niche in a way that facilitates a dense communication network with these teachers to amplify the signals they send towards T cells. Thereby, we will generate a T cell academy that results in a proper training of potent T cells that elicit successful and durable anti-tumor immune responses.